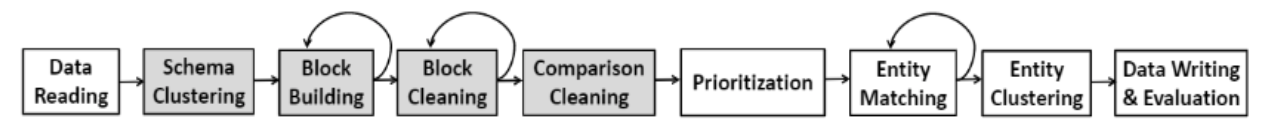
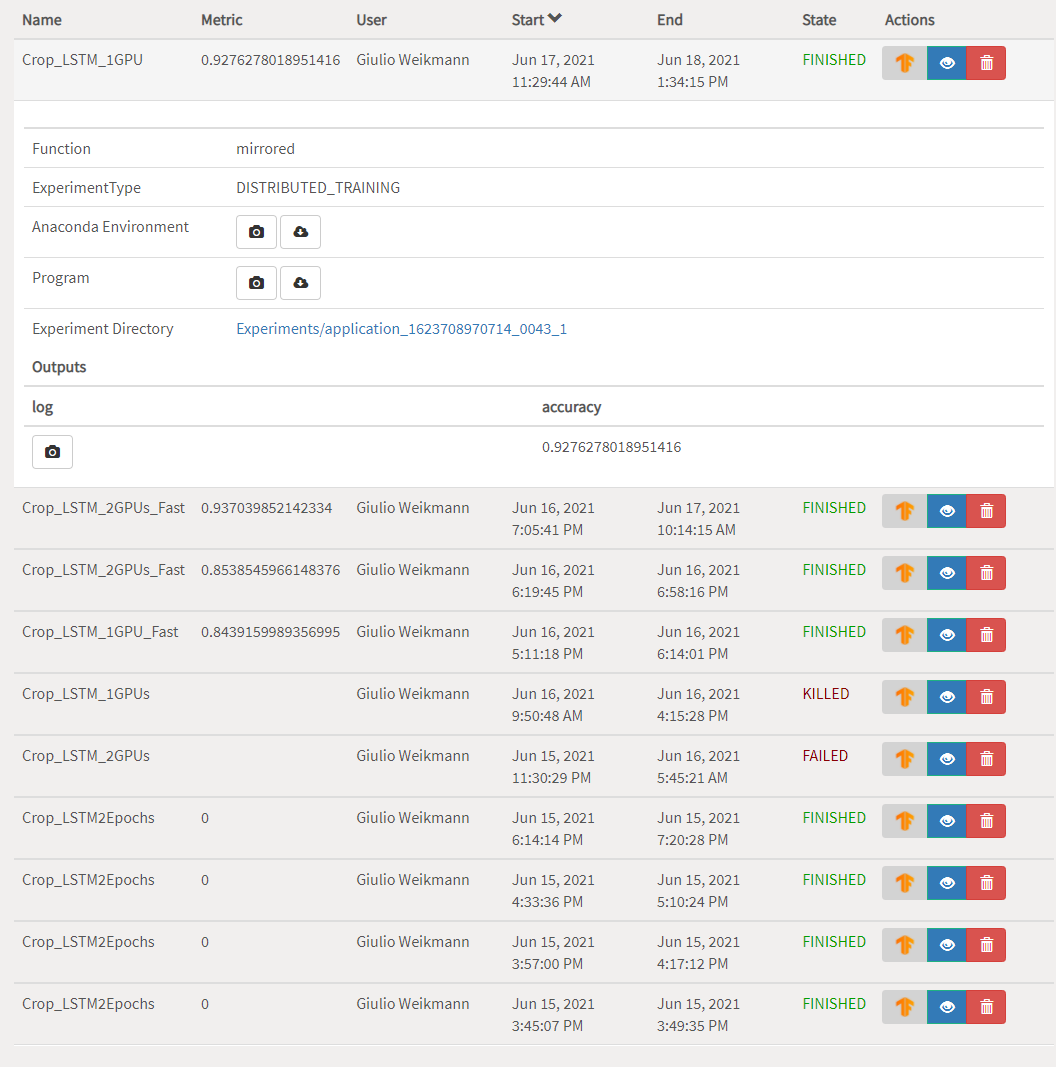
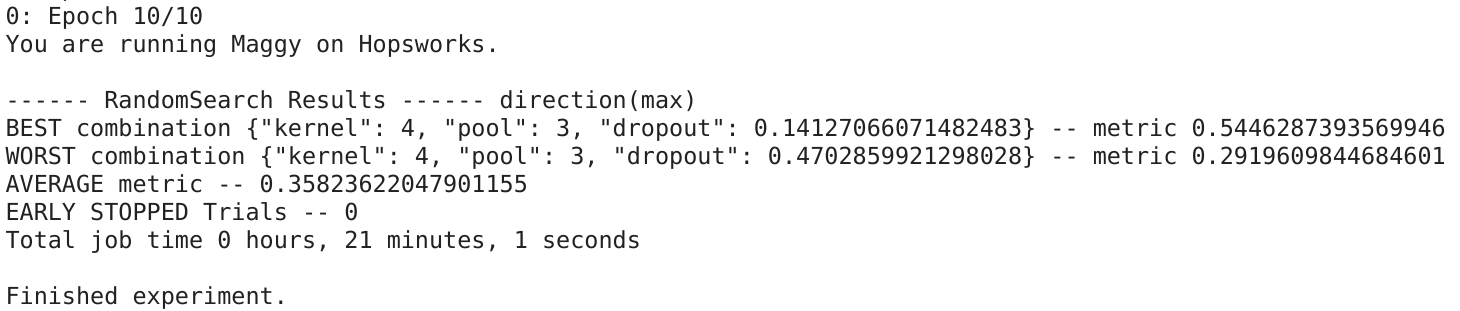
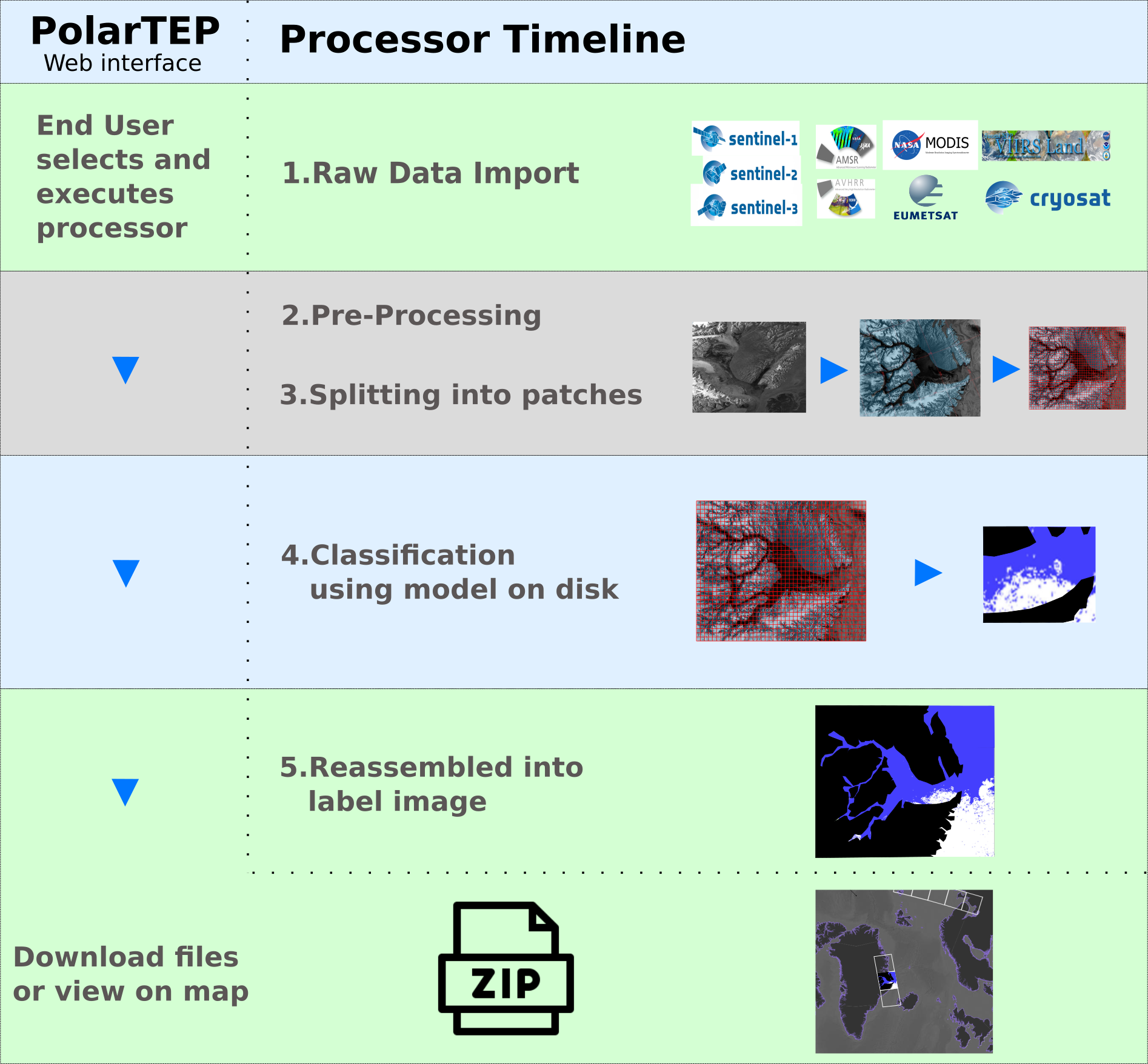
Federating big linked geospatial data sources with Semagrow
Introduction
Semagrow [1] is an open source
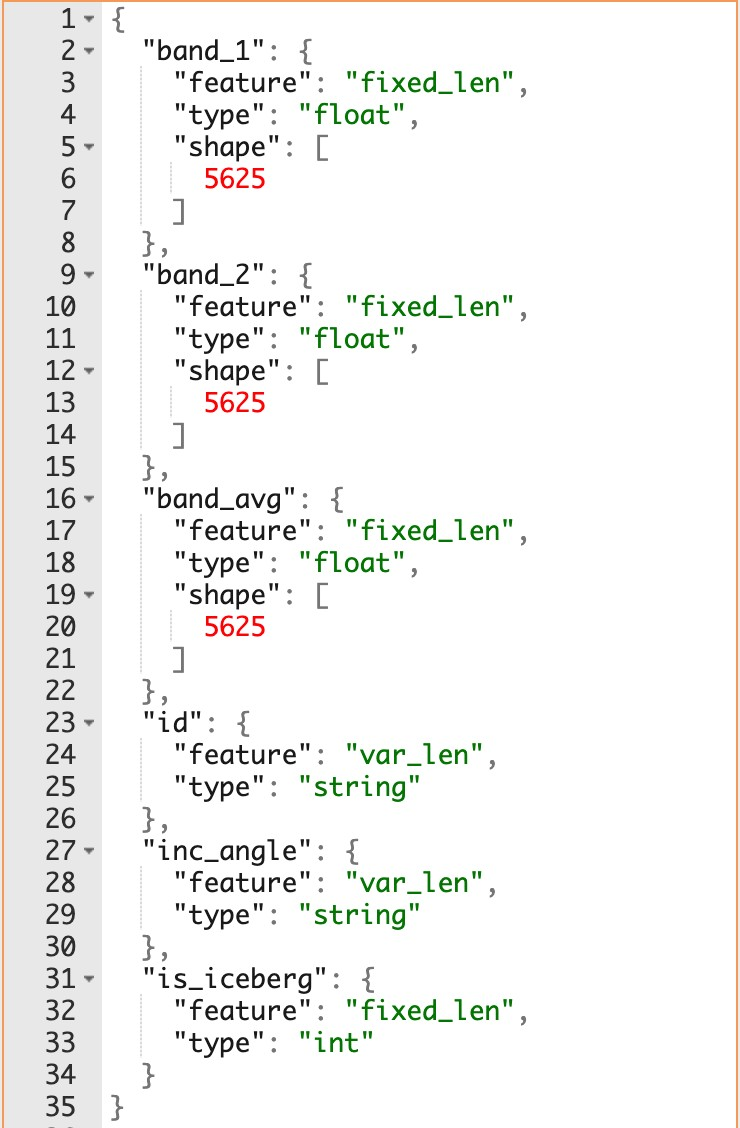
federated SPARQL query processor that allows combining, cross-indexing and, in general, making
the best out of all public data, regardless of their
size, update rate, and schema. Semagrow offers a single SPARQL endpoint that serves data from
remote data sources and that hides from client applications heterogeneity in both form
(federating non-SPARQL endpoints) and meaning (transparently mapping queries and query results
between vocabularies).
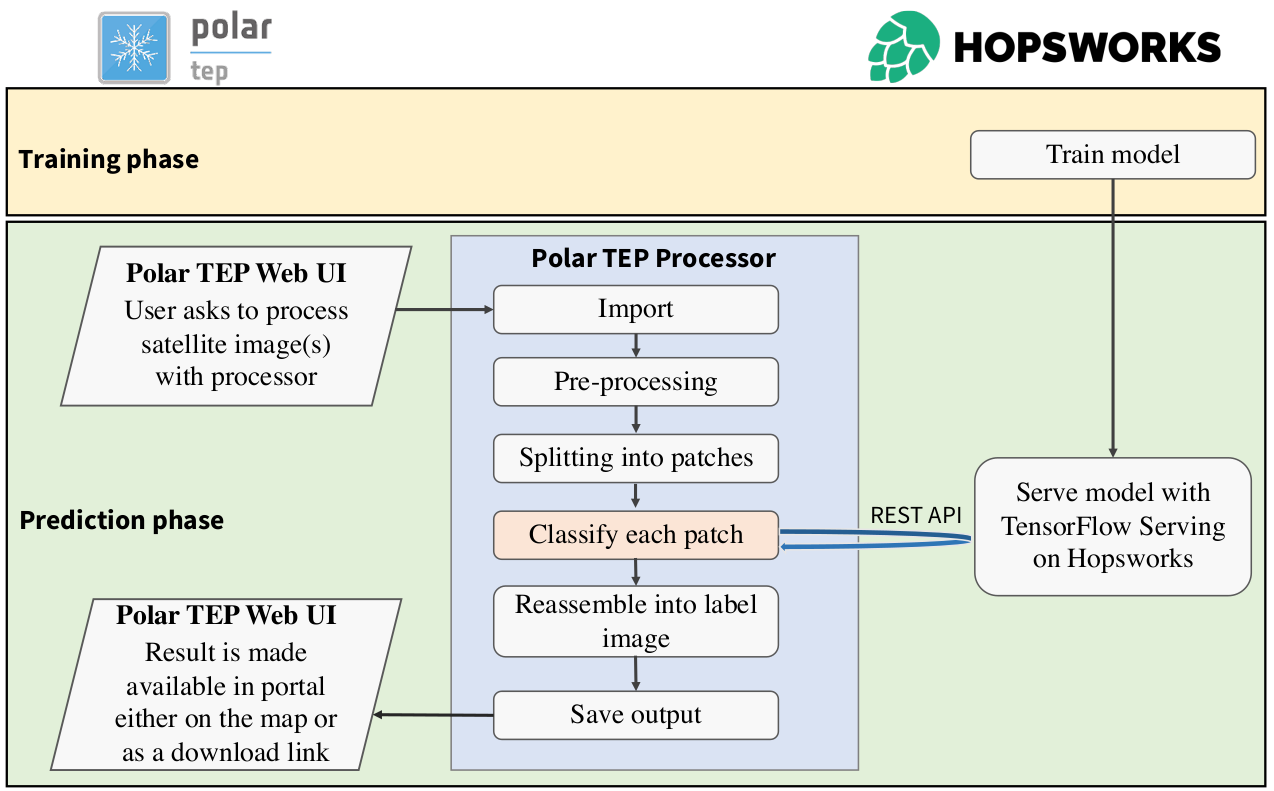
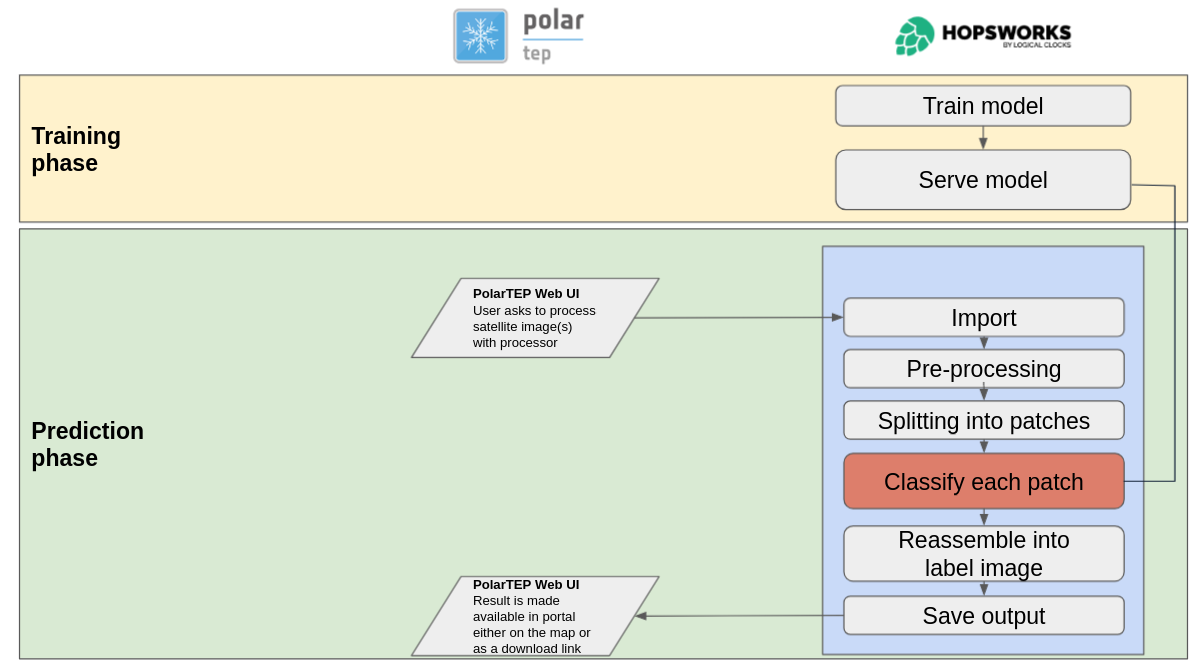
During the Extreme Earth Project, we have developed a new version of Semagrow. The new version
has several extensions and adaptations in order to be able to federate multiple geospatial data
sources, a capability that was, for the most part, missing in the previous version [2], which
was only able to federate one geospatial store with several thematic stores [3]. We have tested
these functionalities in two exercises which use data and queries from the Extreme Earth
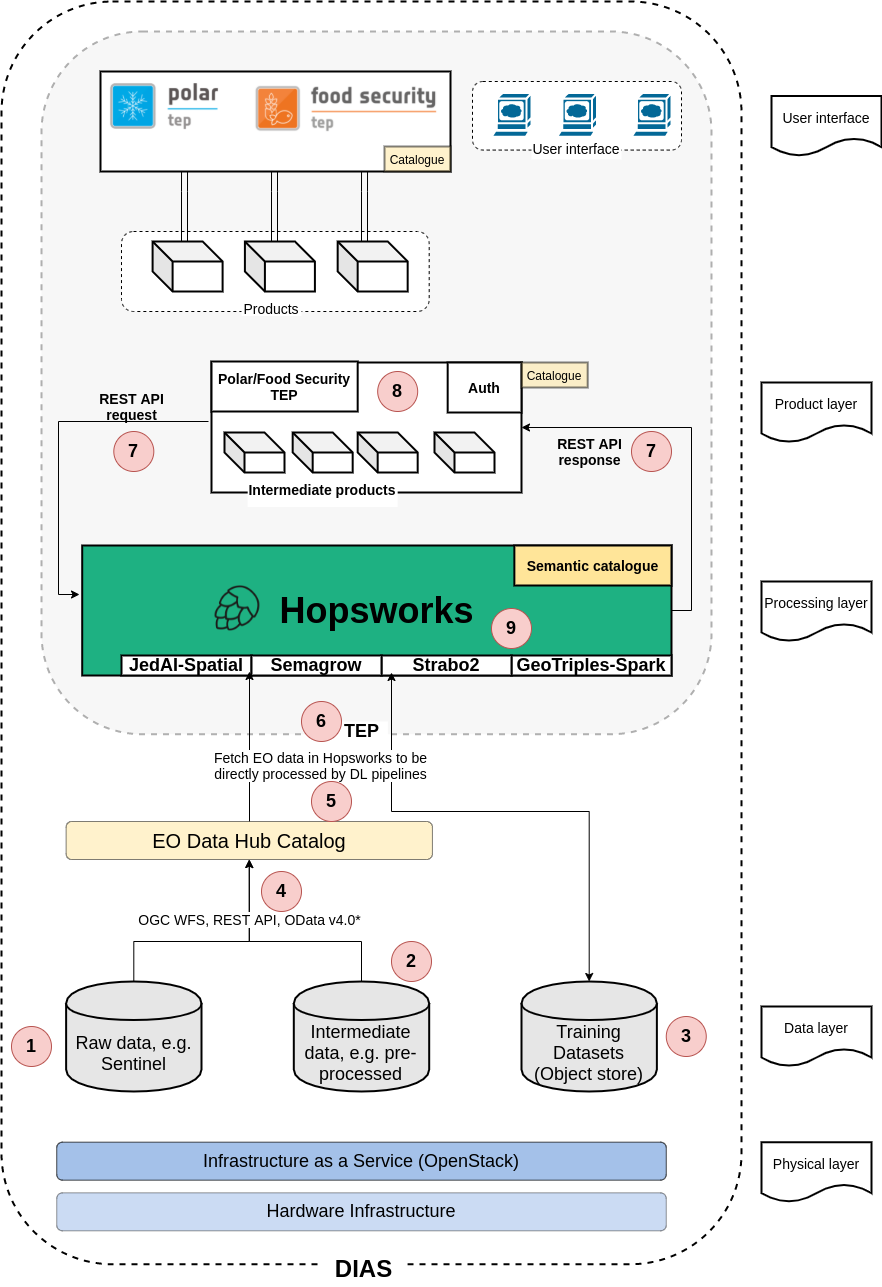
project, and finally, we have integrated Semagrow in Hopsworks as a means for providing a single
access point to multiple data sources deployed in the Hopsworks platform.
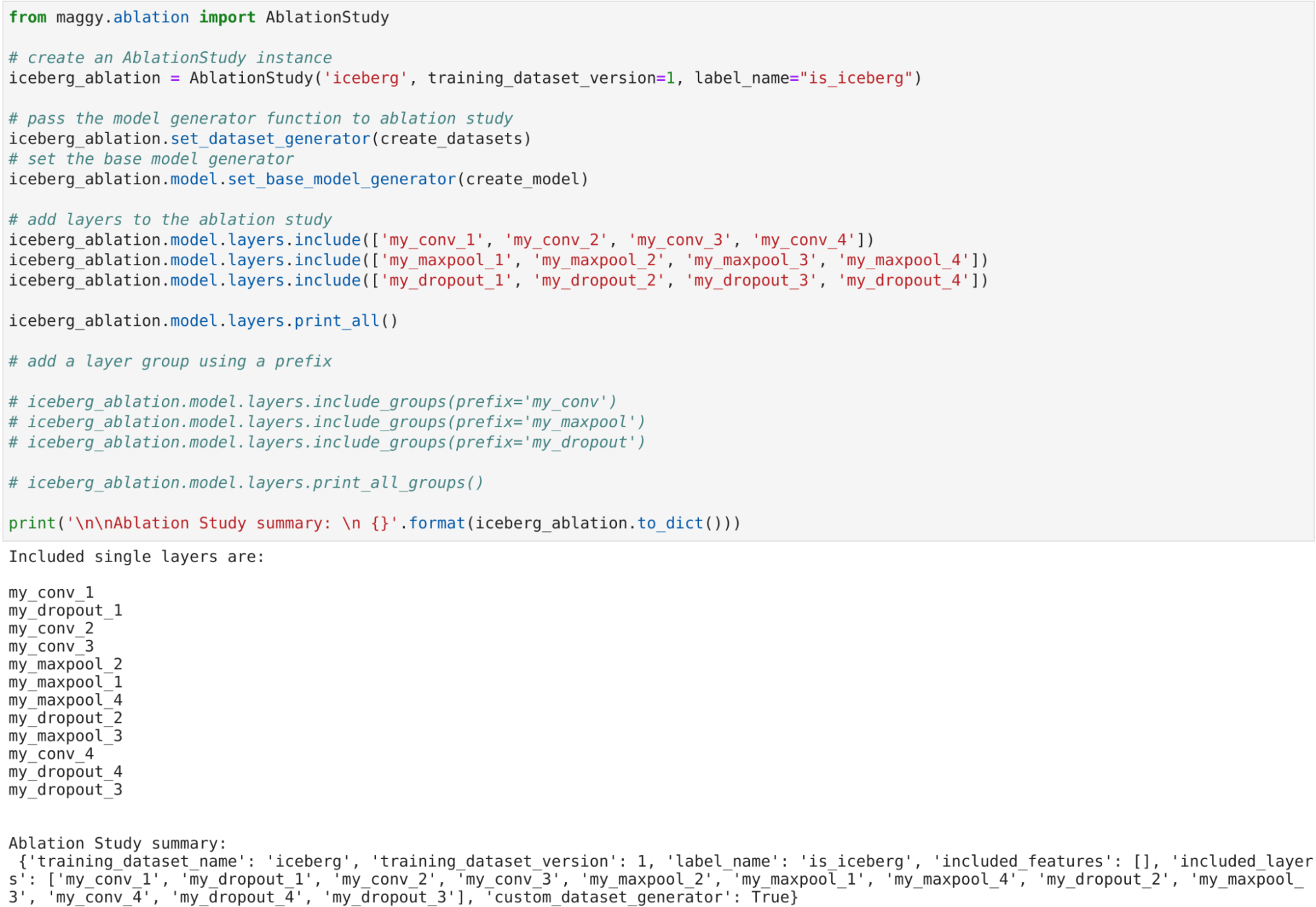
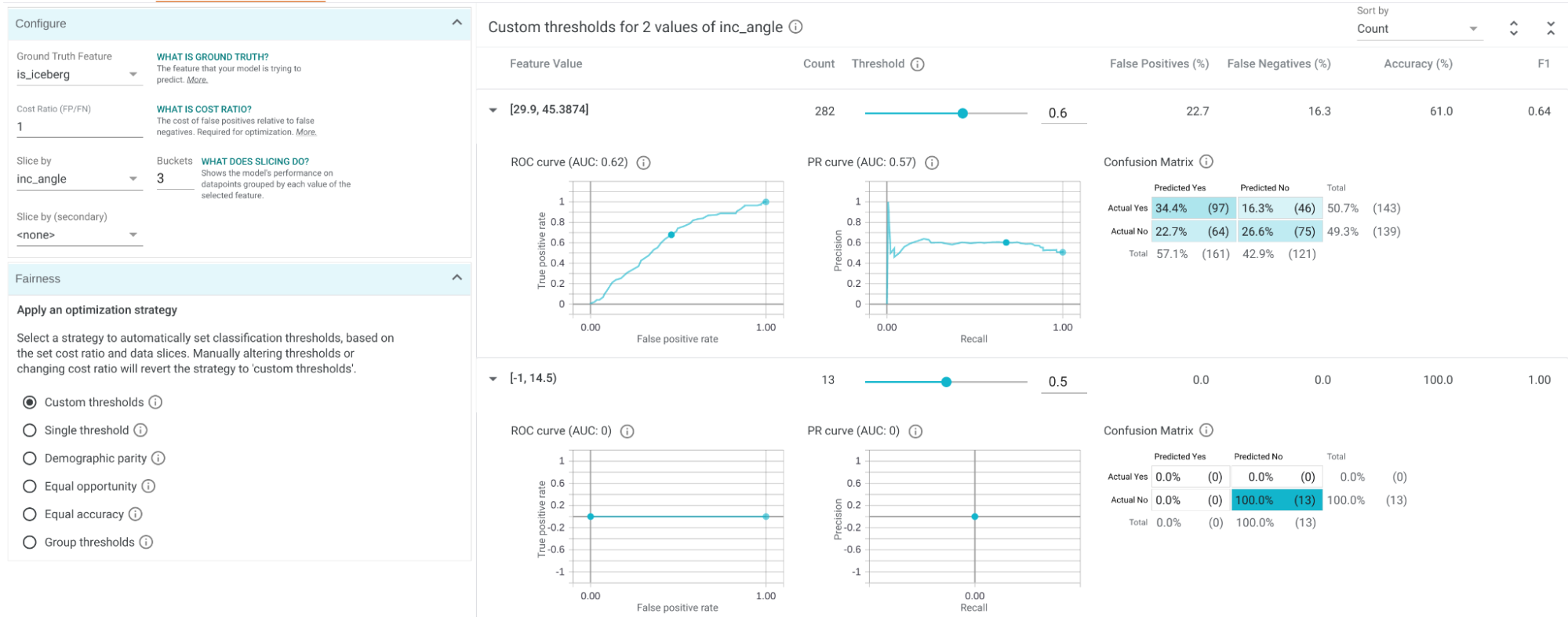
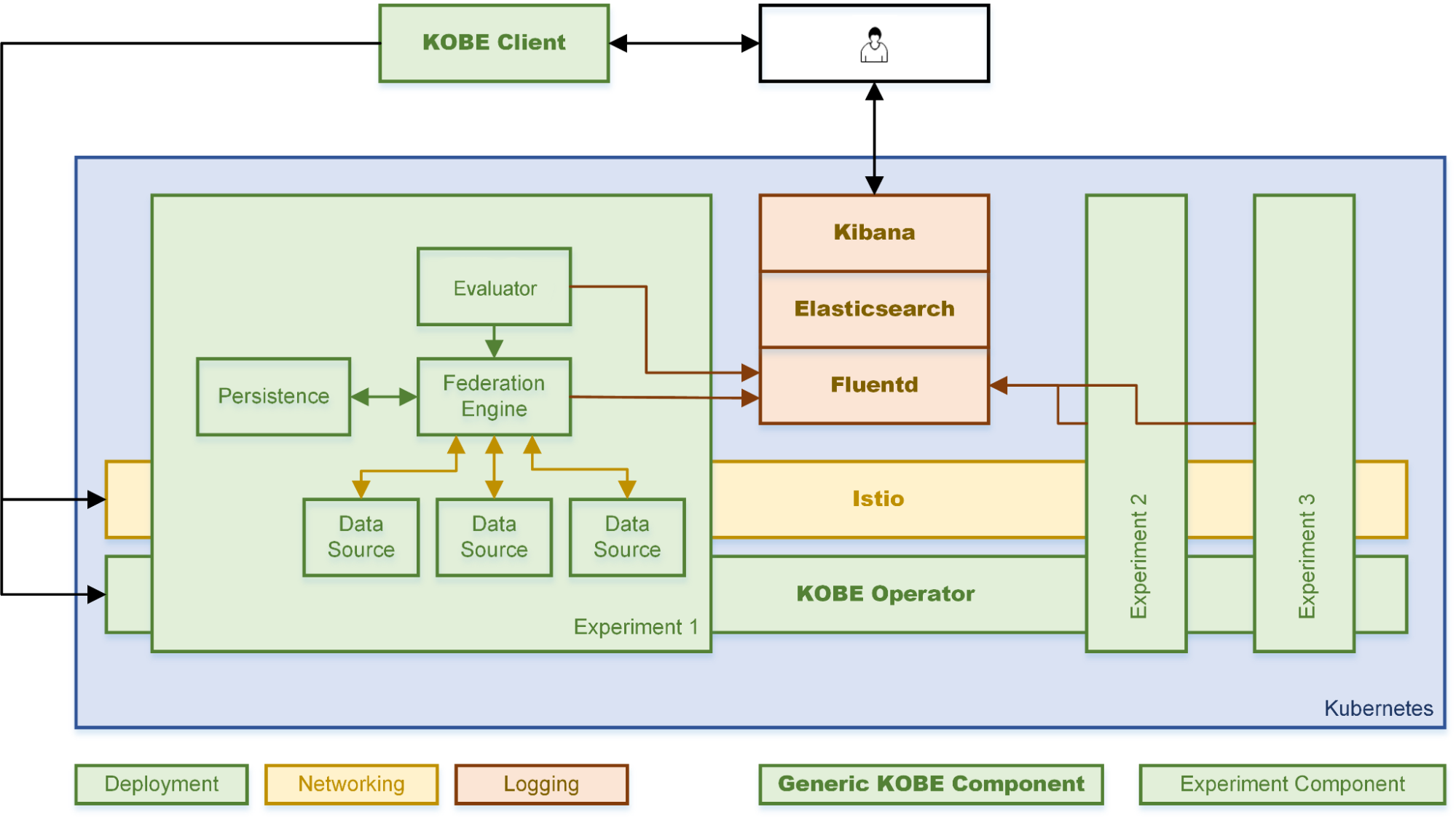
System Description
Semagrow receives a query through its endpoint, decomposes the query into several subqueries,
issues the subqueries to the federated endpoints, combines the results accordingly, and presents
the result to the user. In order to be able to extend Semagrow's capabilities to federated
linked geospatial data, we developed novel techniques for federated geospatial linked data, and
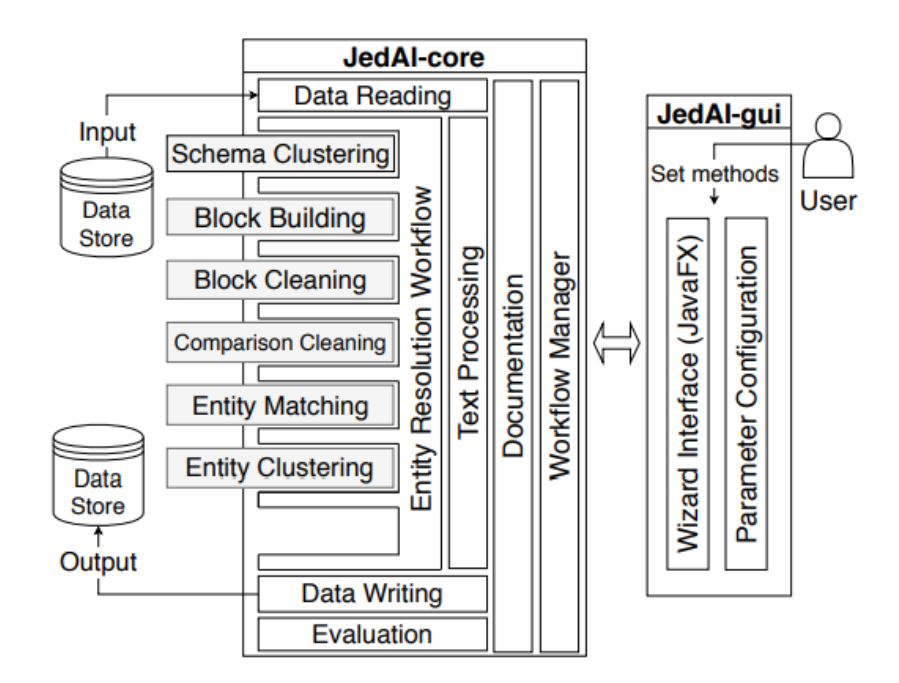
we have extended and re-engineered almost every component of Semagrow's architecture [4]. In the
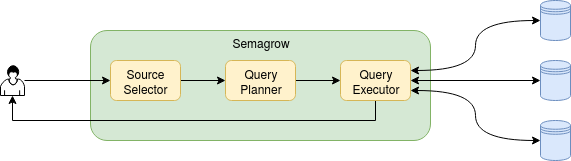
following figure, we illustrate the information flow in Semagrow, and the three major components
that comprise Semagrow’s architecture:
In the remainder of the section, we will describe the current state of Semagrow, by discussing
the improvements to each component in detail.
The first component of Semagrow is the source selector, which identifies which of the federated
sources refer to which parts of the query. In particular, the goal for the source selector is to
exclude as many redundant sources as possible, but without removing any source that contains
necessary data for the evaluation of the query. Semagrow uses a sophisticated source selector
that combines two mechanisms; one that targets thematic data and makes use of all the
state-of-the-art source selection methods in the federated linked data literature; and a novel
geospatial source selection mechanism that targets geospatial linked data specifically [5]. The
idea behind this novel approach is to annotate all federated data sources with a bounding
polygon that summarizes the spatial extent of the resources in each data source, and to use such
a summary as an (additional) source selection criterion in order to reduce the set of sources
that will be tested as potentially holding relevant data. This method can be useful in practice,
because geospatial datasets are likely to be naturally divided in a canonical geographical grid
(consider, for example, weather and climate data) or following administrative regions or, more
generally, areas of responsibility (consider census data as an example).
The second component of Semagrow is the query planner, which uses the result of the source
selector in order to construct an efficient query execution plan by arranging the order of the
subqueries in an optimal way. Semagrow uses a dynamic-programming-based approach and exploits
statistical information about each of the federated sources and a cost model for forming the
optimal plan of a federated query. The effectiveness of the query planner is increased with the
use of additional pushdown optimizations. Pushing the evaluation of some operations in the
federated sources can be effective not only due to the reduction of the communication cost, but
also because geospatial operations are computed faster by the remote GeoSPARQL endpoints; notice
that, in general, a geospatial source maintains its own spatial index for evaluating geospatial
relations. Finally, the query planner of Semagrow is able to process complex GeoSPARQL queries
that were derived from the use cases of Extreme Earth [6]. These queries have complex
characteristics (such as nested subqueries and negation in the form of “not exists” operation),
and, to the best of our knowledge, cannot be processed by any other state-of-the-art federation
engine.
The third component of Semagrow is the query executor, which evaluates the query execution plan,
by providing a mechanism for issuing queries to the remote endpoints and an implementation of
all GeoSPARQL operators that can appear in the plan. Apart from standard GeoSPARQL endpoints,
Semagrow can support several non-SPARQL dataset servers through a series of executor plugins. We
offer a plugin for communicating directly with PostGIS databases with shapefile data that
contain geometric shapes exclusively. For implementing federated geospatial joins, Semagrow uses
bind join with a filter pushdown (i.e., it fetches “left-hand” shapes to partially bind
two-variable functions and pushes the filter to the “right-hand” endpoint). This approach is
effective for standard spatial relations, but not for federated within-distance queries, i.e.,
for queries where the distance between two shapes from different sources has to be less than a
specific distance. This happens because standard spatial relations can be answered very fast by
the spatial indexes of the source endpoints, while the distance operator cannot. For speeding up
within-distance queries, we offer an optimization which rewrites each subquery by inserting
additional geospatial filters that filter out results that are too far away, but these
additional filters use standard spatial relations (and can be executed fast by the source
endpoints).
We mentioned that several parts of Semagrow rely on dataset metadata (e.g., the boundaries of the
source endpoints for the geospatial selector). For this reason, we provide sevod-scraper, which
is a tool that creates dataset metadata [7] from RDF dumps.
Extreme Earth Use Cases
The new version of Semagrow was tested and evaluated using data and queries derived from
practical use cases in the context of Extreme Earth. In particular, we have used Semagrow in the
following use cases: (a) linking land usage data with water availability data provided for the
Food Security Use
Case; and (b) linking land usage data with ground observations for the purpose
of estimating crop type accuracy.
Food security is one of the most challenging issues of this century, mainly due to population
growth, increased food consumption, and climate change. The goal is to minimize the risks of
yield loss while making sure not to damage the available resources. Of great importance is the
study of irrigation, which requires reliable water resources in the area being farmed.
Considering the fact that a large portion of the world's freshwater is linked to snowfall and
snow storage, a promising way for providing an indication of water availability for irrigation
is to study the snow cover areas in conjunction with the field boundaries and their crop-type
information. The queries that are most relevant for this analysis are spatial within queries,
spatial intersection queries, and within-distance queries: that is, retrieving the land parcels
with a given crop type that are within, intersecting, or within a given maximum distance
(without requiring the exact distance) from any snow-covered area. Moreover, sometimes we need
to reduce our focus either on a smaller polygonal area with given coordinates, or on a specific
administrative region.
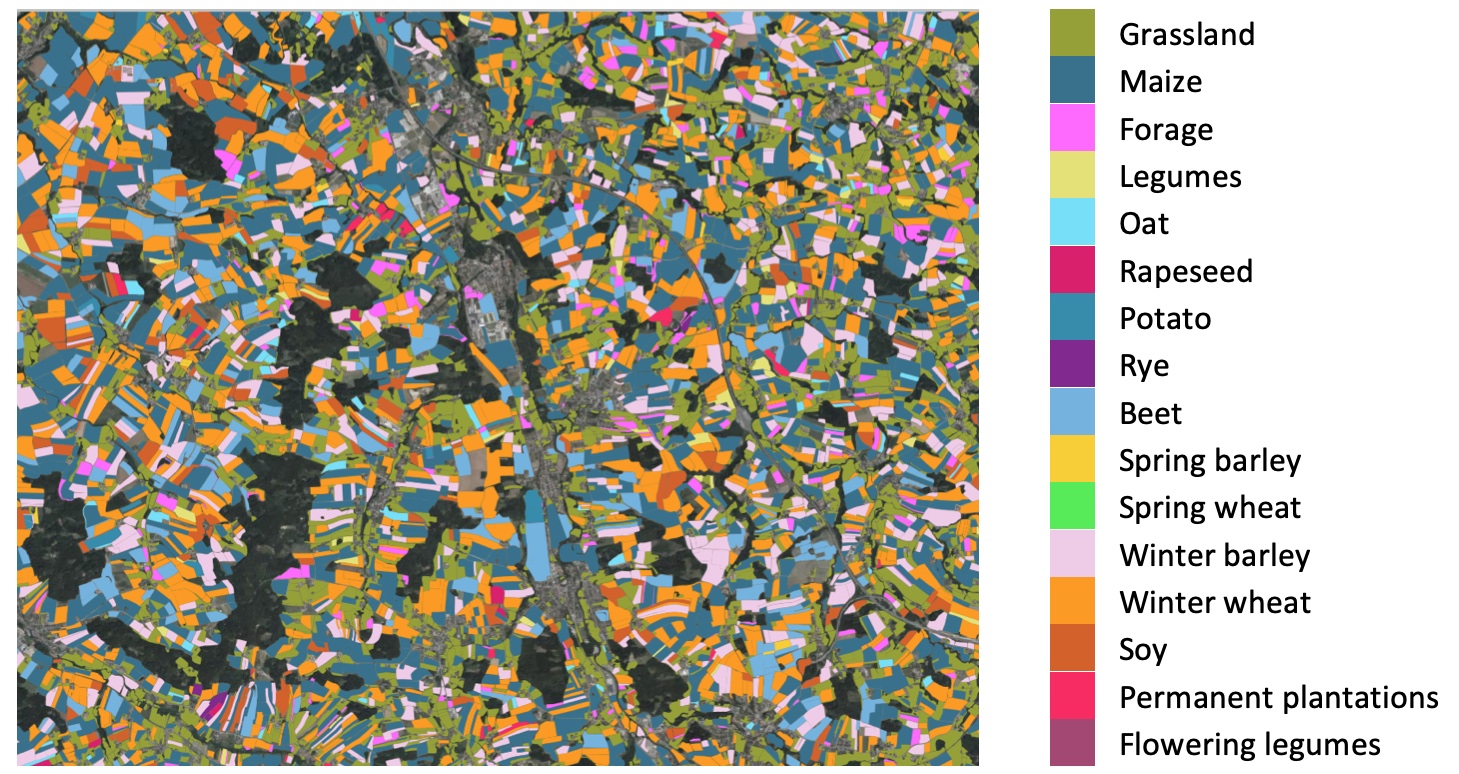
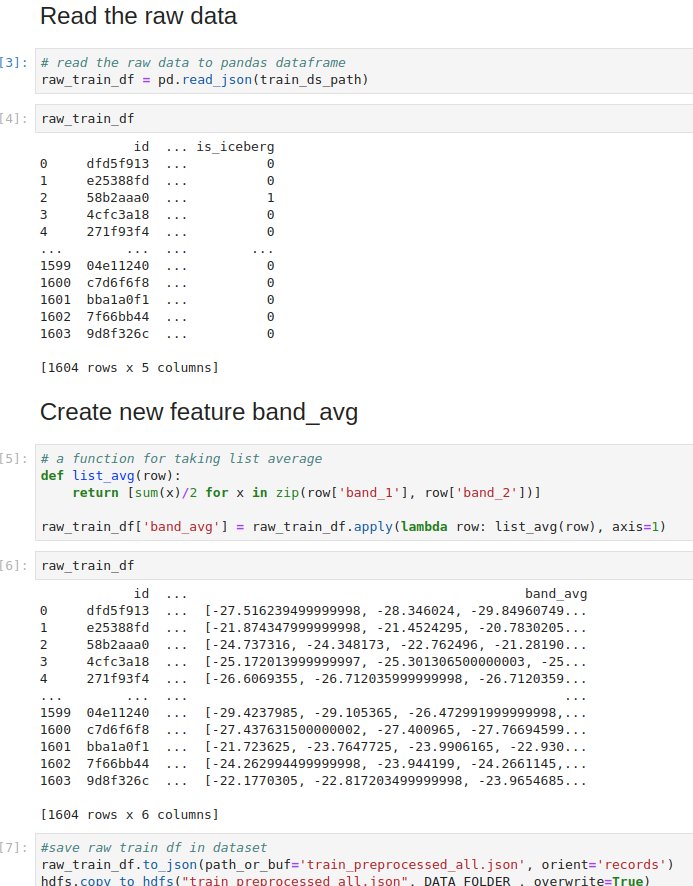
We have used Semagrow in an exercise based on the above use case as follows: We created a
federation that consists of geospatial RDF data that cover Austria in three three data layers
(i.e., snow, crop, and administrative data). In particular, we envisage that Austrian state
governments publish crop datasets for their own area of responsibility; and a further
(different) entity publishes a snow cover dataset that ignores state boundaries and publishes
its datasets according to a geographical grid. Therefore, each federated endpoint contains data
from a specific data layer and for a specific area (e.g., crop data of the state of Upper
Austria). We used Strabo2 for serving the data. Since Semagrow is a standard GeoSPARQL endpoint,
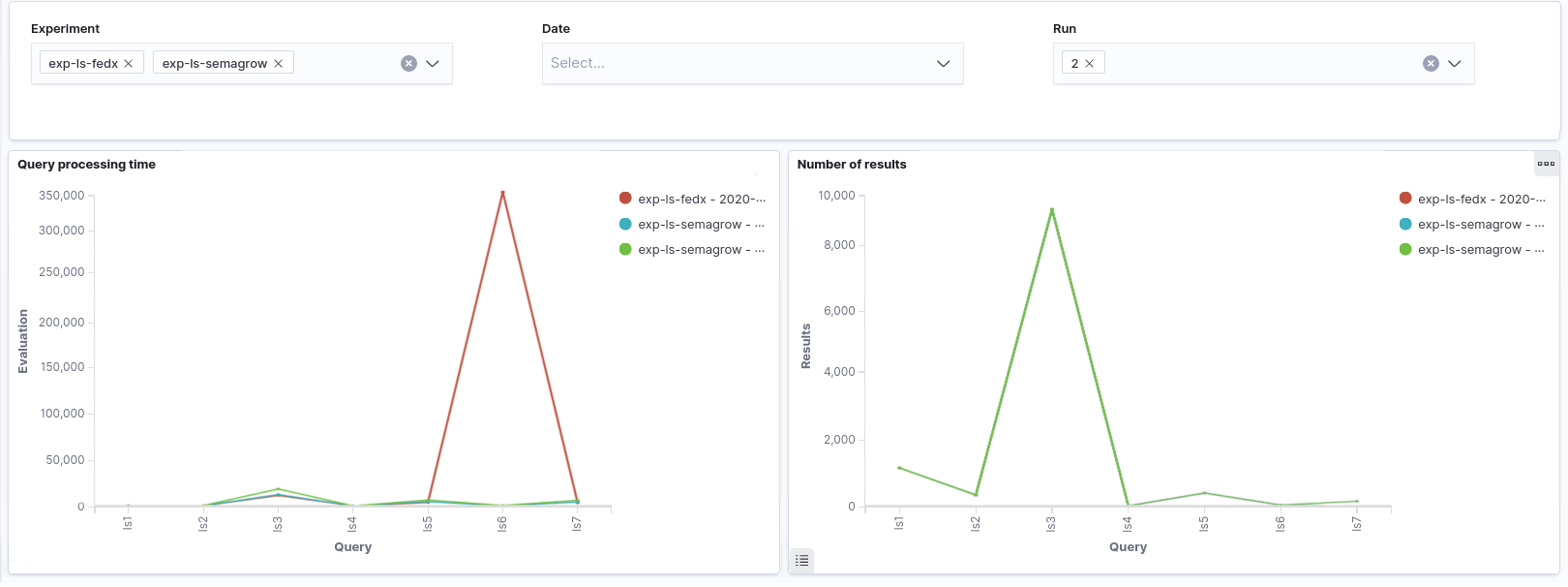
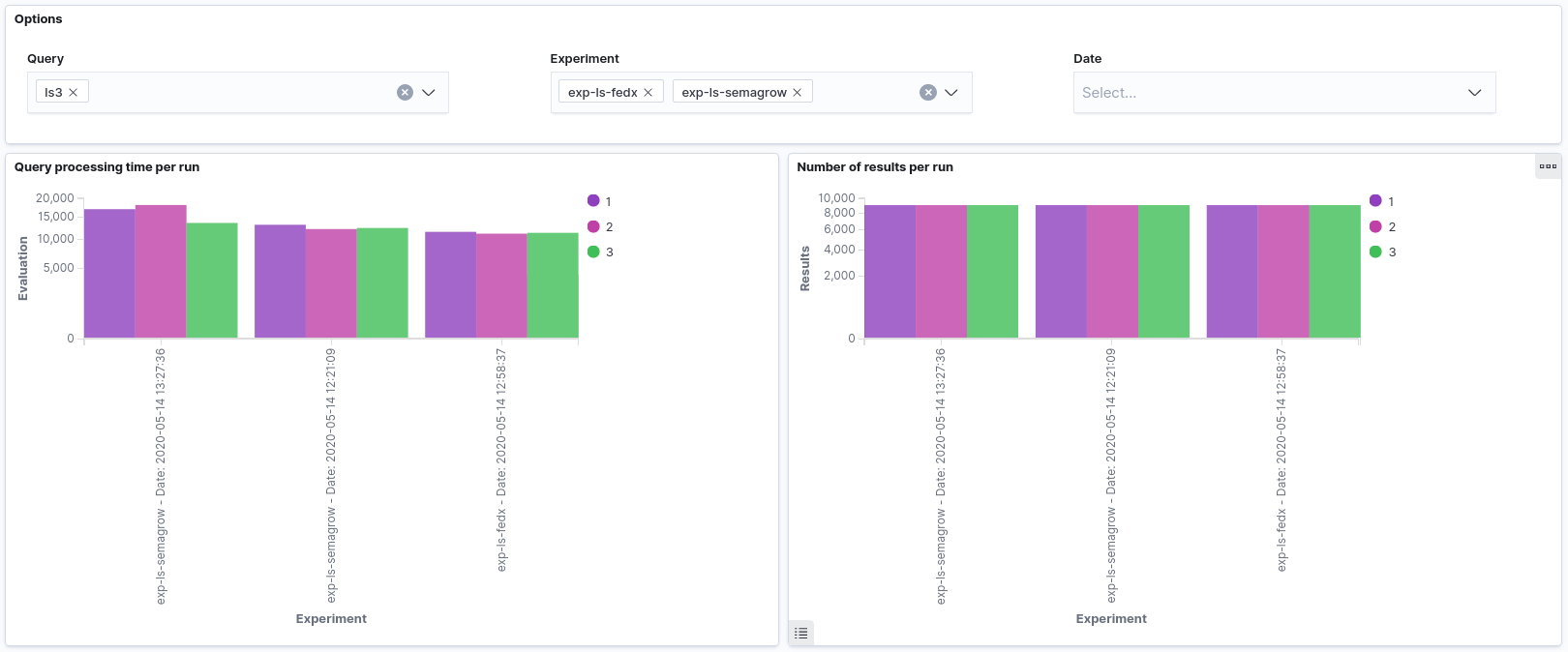
the results were able to be visualized in Sextant. In this series of experiments, we observed
that the new version of Semagrow performs better than the old one, mainly due its source
selector. Since the new source selector is aware both of the geospatial and thematic nature of
the federated sources, it is able to effectively reduce the sources that appear in the query
execution plan, and as a result to reduce the source queries issued by the query executor to the
endpoints of the federation.
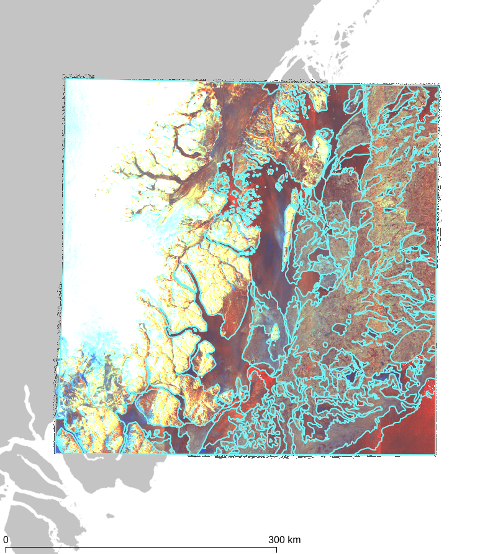
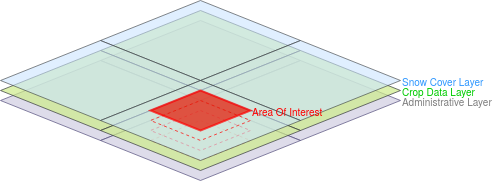
In the following figure, we illustrate a simplified version (4 snow, 4 crop, and 4 administrative
sources, non-overlapping and perfectly aligned bounding polygons) of the above case. Assume that
we want to retrieve all snow-covered crops within a specific area of interest shown in red. In
this case, our source selection first prunes all administrative sources since they contain
irrelevant thematic data; and then it prunes the sources that are disjoint from the area of
interest since they contain irrelevant geospatial data. As a result, the query will be evaluated
faster since the query execution plan will contain only 2 (of the total 12) sources.
Detailed land usage data is crucial in many applications, ranging from formulating agricultural
policy and monitoring its execution, to conducting research on climate change resilience and
future food security. Land usage can be inferred from Earth Observation images or collected
through self-declaration, but in either case it is important for such data to be validated
against land surveys. Ground observations are geo-referenced to a point on the road adjacent to
a field (and not inside a field), which is often ambiguous in agricultural areas with several
adjacent parcels; further exacerbated by GPS accuracy. Therefore, we can estimate the error rate
of the land usage data as follows: first, all ground observation is irrelevant to the analysis
if it is more than 10 meters from any crop parcel. For the remaining ground observations, we
find the nearest land parcel, and if the crop types match, the GPS point provides a positive
validation; otherwise it provides a negative one. This process is challenging not only because
it is computationally demanding (since it involves quadratic many distance calculations), but
because the crop types between different datasets usually make use of different code names.
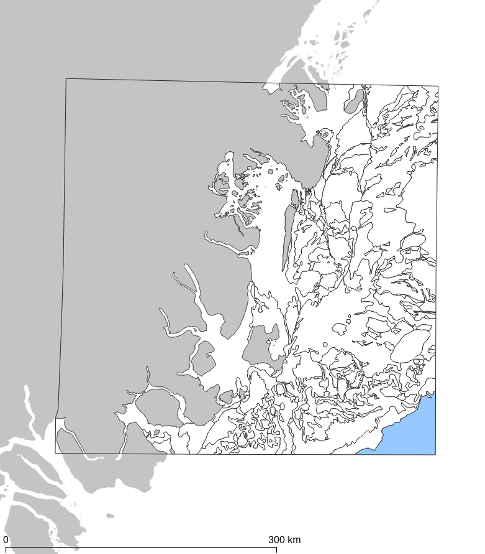
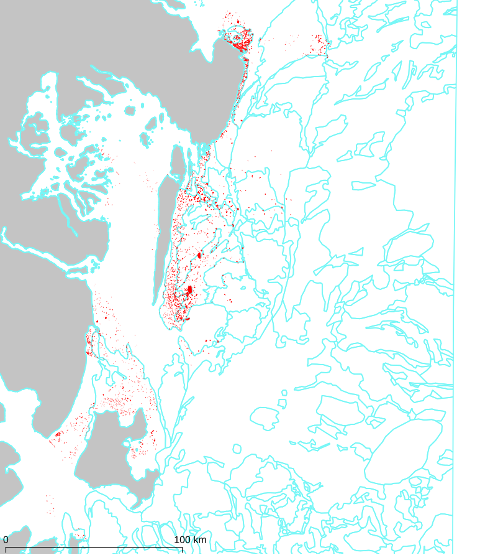
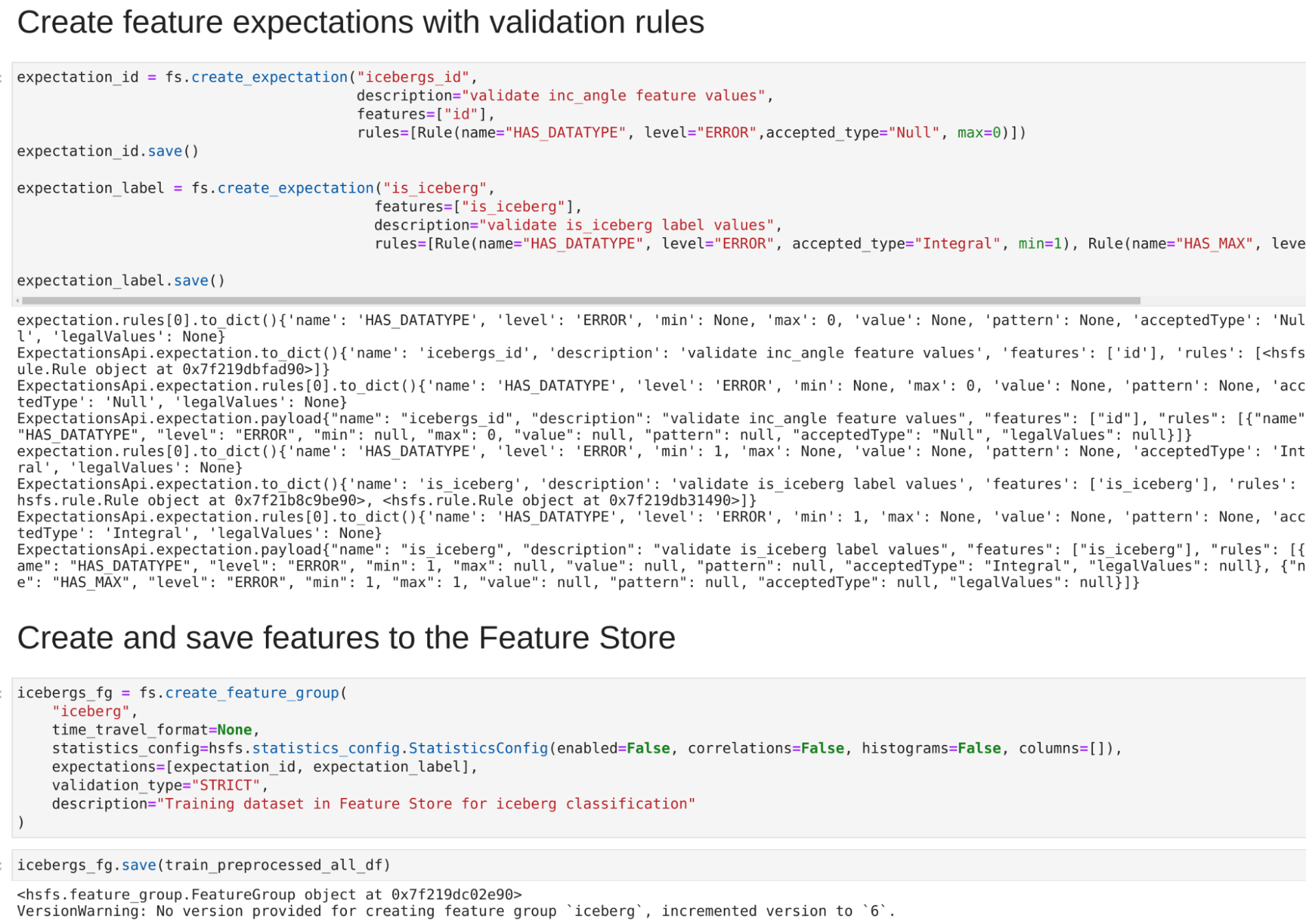
In the following figure, we illustrate 3 ground observations located in the roads adjacent to
field parcels, used for crop-type validation of the field dataset. Notice that two of them (the
green ones) provide a positive and the other one provides a negative validation.
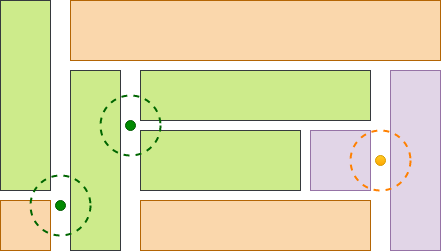
We have used Semagrow for the task of the crop-data validation of the Austrian Land Parcel
Identification System (INVEKOS), which contains the geo-locations of all crop parcels in
Austria
and the owners' self-declaration about the crops grown in each parcel. This dataset was
validated using the EUROSTAT's Land Use and Cover Area frame Survey (LUCAS), which
contains
agro-environmental and soil data by field observation of geographically referenced points. Both
datasets were transformed into RDF with GeoTriples, and were deployed in two separate Strabo2
endpoints. The new version of Semagrow completes the task very efficiently due to the federated
within-distance optimizer; even though the queries of the task contain several complex
characteristics (such as subqueries and negation), the bottleneck of the evaluation is the
calculation of the within-distance operator. By disabling the optimization, the processing of
the queryload would require several days, while with the optimization in place, the task reduces
to several hours.
Conclusion
In this post we presented the new version of the Semagrow query federation engine, which was
developed during the Extreme Earth project. Semagrow provides unified access to big linked
geospatial data from multiple, possibly heterogeneous, geospatial data servers. All phases of
the federated query processing (namely source selection, query planning and query execution) of
Semagrow were extended, and Semagrow has been successfully integrated within Hopsworks. This new
version of Semagrow was used and evaluated on data and queries from the use cases of the Extreme
Earth project.
References
[1] Angelos Charalambidis, Antonis Troumpoukis, Stasinos Konstantopoulos: SemaGrow: optimizing
federated SPARQL queries. In SEMANTICS 2015, Vienna, Austria, September 15-17, 2015
[2] Stasinos Konstantopoulos, Angelos Charalambidis, Giannis Mouchakis, Antonis Troumpoukis,
Jürgen Jakobitsch, Vangelis Karkaletsis: Semantic Web Technologies and Big Data Infrastructures:
SPARQL Federated Querying of Heterogeneous Big Data Stores. In ISWC 2016 (Posters & Demos),
Kobe, Japan, October 17-21, 2016
[3] Athanasios Davvetas, Iraklis Klampanos, Spyros Andronopoulos, Giannis Mouchakis, Stasinos
Konstantopoulos, Andreas Ikonomopoulos, Vangelis Karkaletsis: Big Data Processing and Semantic
Web Technologies for Decision Making in Hazardous Substance Dispersion Emergencies. In ISWC 2017
(Posters, Demos & Industry Tracks), Vienna, Austria, October 21-57, 2017
[4] Antonis Troumpoukis, Nefeli Prokopaki-Kostopoulou, Giannis Mouchakis, Stasinos
Konstantopoulos: Software for federating big linked geospatial data sources, version 2, Public
Deliverable D3.8, Extreme Earth Project, 2021.
[5] Antonis Troumpoukis, Nefeli Prokopaki-Kostopoulou, Stasinos Konstantopoulos: A Geospatial
Source Selector for Federated GeoSPARQL querying. Article in Preparation.
[6] Antonis Troumpoukis, Stasinos Konstantopoulos, Giannis Mouchakis, Nefeli
Prokopaki-Kostopoulou, Claudia Paris, Lorenzo Bruzzone, Despina-Athanasia Pantazi, Manolis
Koubarakis: GeoFedBench: A Benchmark for Federated GeoSPARQL Query Processors. In ISWC
(Demos/Industry) 2020, Online, November 1-6, 2020
[7] Stasinos Konstantopoulos, Angelos Charalambidis, Antonis Troumpoukis, Giannis Mouchakis,
Vangelis Karkaletsis: The Sevod Vocabulary for Dataset Descriptions for Federated Querying. In
PROFILES@ISWC 2017, Vienna, Austria, October 21-57, 2017